| Table of Contents |
|---|
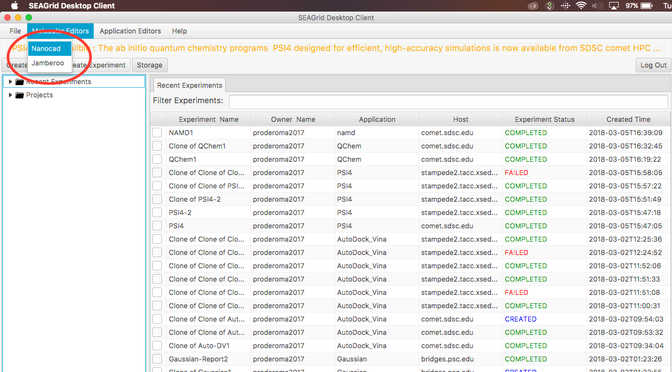
- Login to SEAGrid desktop client.
- Select Nanocad editor
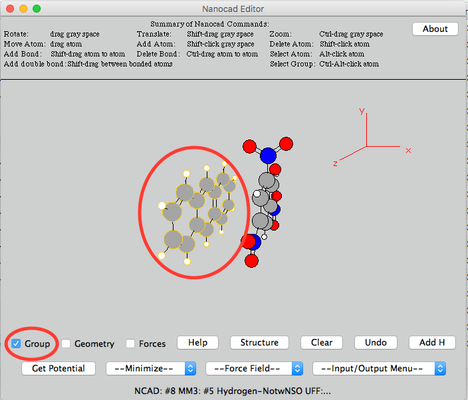
Image I - Desktop client home. Select Nanocad - After NanoCAD has launched, take your time to read text "Summary of NanoCAD Commands" at the top of NanoCAD window (Indicated by the green box in Image I).
- You will get basic idea how to use NanoCAD
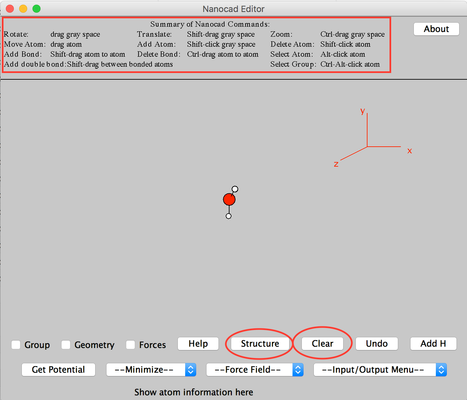
- Click 'Clear' to remove the existing water molecule.
- Click 'Structure'.
Image II - Nanocad editor, clear the water molecue
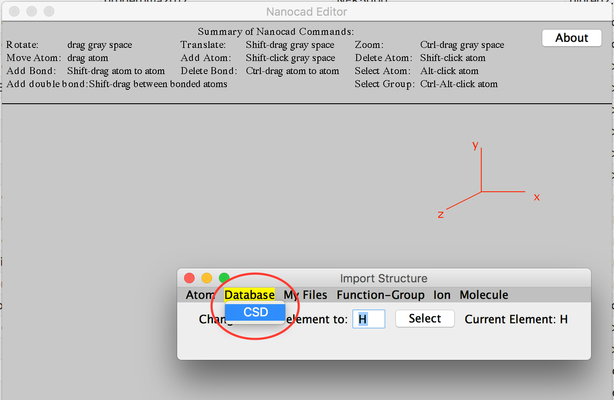
- From 'Import Structure' click Database and then select CSD.
Image III - Select CSD to search for new molecule -
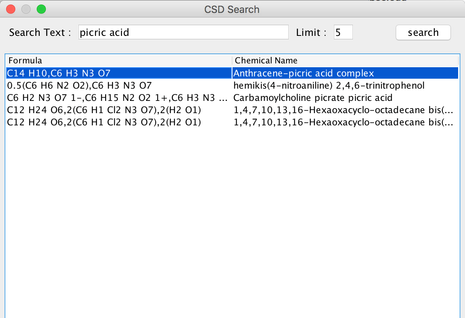
Search for ‘picric acid’ (This would take about 20 seconds). From the list given, double click on the first one ‘anthracene-picric acid complex’.Anchor anchor2 anchor2
Image IV - Picric acid search results in CSD search - The complex will get ported in to the nanocad editor. Select ‘Group’ and do ctrl+alt+click on one of the anthracene atoms. Then once the anthracene group is highlighted, click ‘Clear’ and remove the anthracene molecule
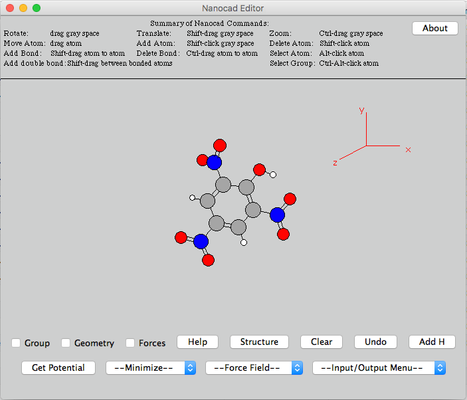
Image V - Anthracene-picric acid complex in nanocad editor - Ctrl+click on the gray background area to center the picric acid molecule. You can rotate the molecule by click and drag.
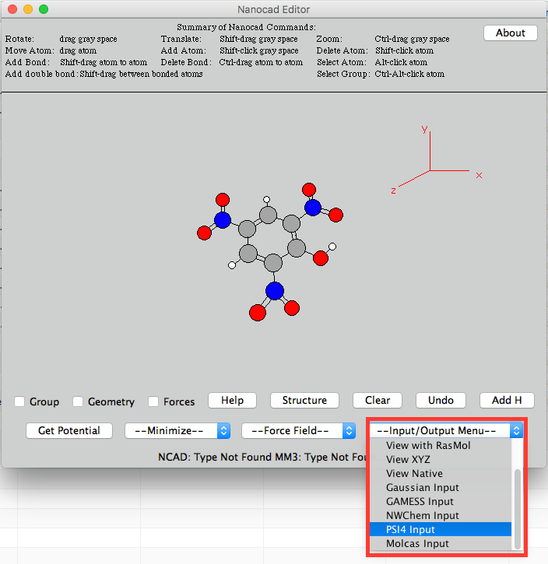
Image VI - Picric acid - Click 'PSI4 Input' from --Input/Output Menu – on lower righthand corner.
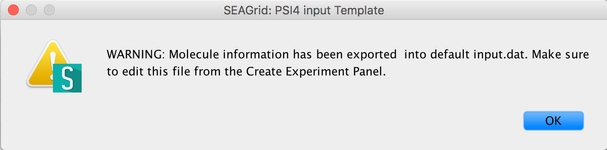
Image VII - Export as a PSI4 Input - Read and say OK, to the warning mesage.
Image VIII - Warning message on exporting molecule.
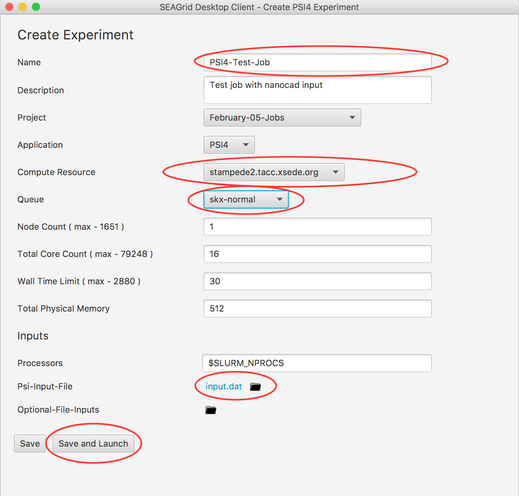
Above input file is then exported in to 'Create a PSI4 Experiment'. You can edit the exported input.dat file, simply click and edit the file and save. If no changes provide experiment name and select the HPC from the available and launch the job.Anchor anchor1 anchor1
Image IX - Creating PSI4 experiment to launch job
...
- Steps are very similar to above PSI4 input build.
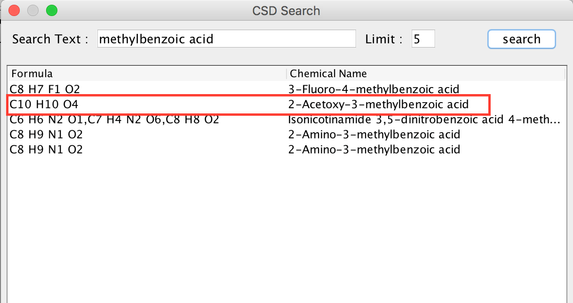
- Follow the first 4 steps similar to 'Build PSI4 Input' and in step 5, search for 'methylbenzoic acid'. From the list select '2-Acetoxy-3-methylbenzoic acid'.
Image I - Selecting 2-Acetoxy-3-methylbenzoic acid from databse - Remove the unwanted groups, in this scenario its the acetoxy group. In order to remove you do a ctrl+ drag from the carbon to oxegeon bond.
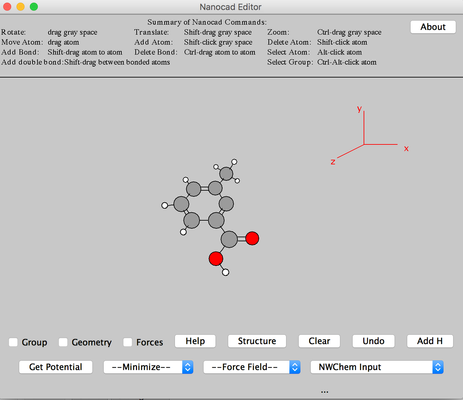
Image II - Removing unwanted acetoxy group - Once the acetoxy group is removed only the methylbenzoic acid will be remaind. Center the molecule by ctrl+click on the gray space.
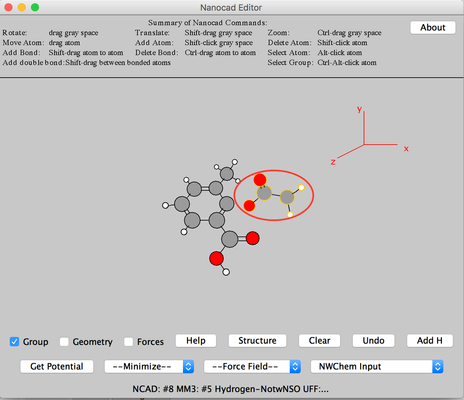
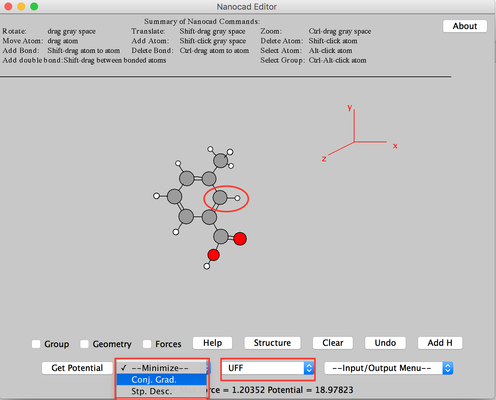
Image III - Center the molecule. - Click Add H. H will be added to the carbon atom. Select 'UFF' from 'Force Field' and 'Conj. Grad' from 'Minimize'.
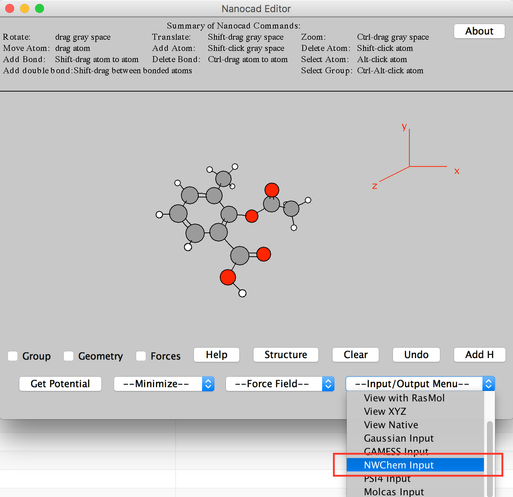
Image IV - Adding H and change of properties - From 'Input/Output Menu' select NWChem Input. Say 'OK' to the warning message.
Image V - Import the molecule - Provide the required fields as per 'step 9' in 'Build PSI4 Input' and launch the experiment.
- Prior to launching the experimnet you could view the uploaded input file and change the content and save. upon saving the file you can launch the experiment with an updated input.
Build Molcas Input
- Steps are very similar to above PSI4 input build.
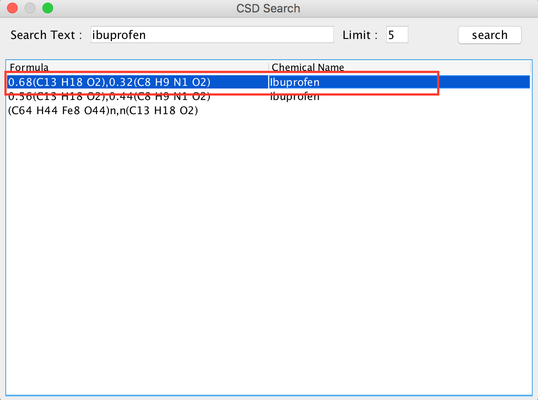
- Follow the first 4 steps similar to 'Build PSI4 Input' and in step 5, search for 'ibuprofen'. From the list select 'ibuprofen'. 335px
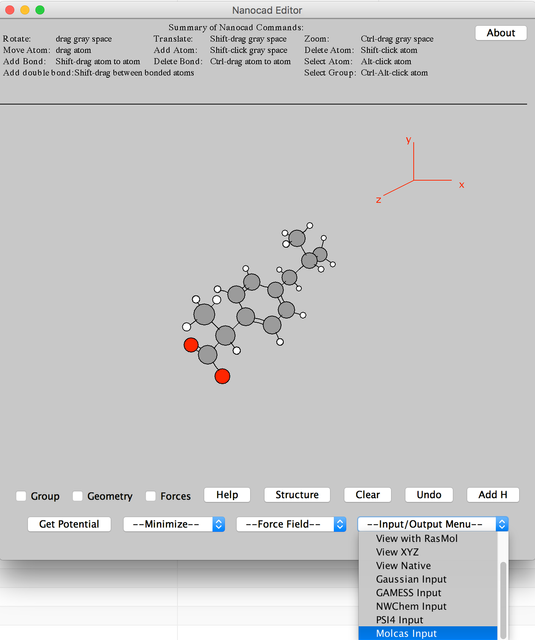
Image I - Select Ibuprofen - In Nanocad editor export the molecule as a Molcas input.
Image II - Export to Create experiment - Provide the required fields as per 'step 9' in 'Build PSI4 Input' and launch the experiment.